Next: Source localization Up: Independent Component Analysis For Previous: Inverse Problem
Statistical preprocessing of the data
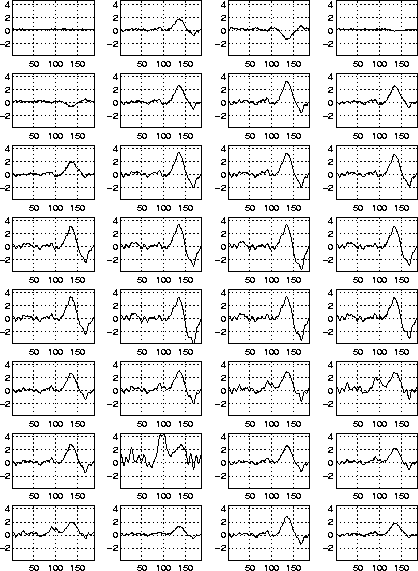
Figure: Simulated scalp potentials due to three dipole sources mapped onto 32 channels (electrodes). Channels are numbered left to right, top to bottom. The first channel is the reference electrode. These signals are the input data for the ICA algorithm. The locations of these 32 electrodes are shown in Fig. 3.
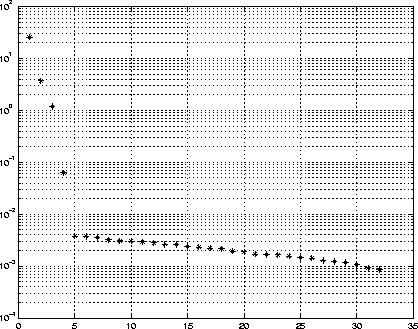
Figure 7: Singular values of the covariance matrix. It appears
that only the first four singular values contribute to the signal subspace,
with the rest constituting the noise subspace.
In EEG experiments, electric potential is measured with an array of electrodes (typically 32, 64, or 128) positioned primarily on the top half of the head, as shown in Fig. 3. The data are typically sampled every millisecond during an interval of interest.
For a given electrode configuration, the time-dependent data can be
arranged as a matrix, where every column corresponds to the sampled time
frame and every row corresponds to a channel (electrode). For example,
the data obtained by 32 electrodes in 180 ms can be sampled in 180 frames
and represented as a matrix (32 ![]() 180). Below we will refer to this matrix as
180). Below we will refer to this matrix as ![]() , where instead of a continuous variable t, we have sampled time
frames
, where instead of a continuous variable t, we have sampled time
frames ![]() .
.
Before performing source localization, we will preprocess the EEG activation maps in order to decompose them into several independent activation maps. The source for each activation map will then be localized independently. This is accomplished as follows:
- -
-
First, we will process the raw signals,
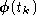 , in order to reduce the dimensionality of the data, and to remove some
of its noise. The projection of the data on the signal subspace will be
referred to as
, in order to reduce the dimensionality of the data, and to remove some
of its noise. The projection of the data on the signal subspace will be
referred to as 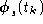 .
. - -
-
The signal subspace,
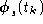 , will then be decomposed into statistically independent terms,
, will then be decomposed into statistically independent terms, 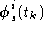 .
. - -
-
Each independent activation,
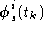 , will be assumed to be due to a single stationary dipole, which we will
then localize using a parameterized search algorithm.
, will be assumed to be due to a single stationary dipole, which we will
then localize using a parameterized search algorithm.
![]()
and constructing signal and noise subspaces [15]. The noise subspace will constitute the singular vectors with singular values less than a chosen noise threshold.
![]()
Having constructed the subspaces, we can project the original data onto the signal subspace by:
where ![]() and
and ![]() are the signal subspace singular values and singular vectors.
are the signal subspace singular values and singular vectors.
Though PCA allows us to estimate the number of dipoles, in the presence
of noise it does not necessarily give an accurate result [15].
In order to separate out any remaining noise, as well as each statistically
independent term, we will use the recently derived infomax technique,
Independent Component Analysis (ICA). (It is worth noting that PCA not
only filters out noise from the data, but also makes a preliminary step
of ICA decomposition by decorrelating the channels, or removing linear
dependence, i.e., ![]() . ICA then makes the channels independent, i.e.,
. ICA then makes the channels independent, i.e., ![]() for any powers n and m.)
for any powers n and m.)
There are several assumptions one needs to make about the sources in order to apply the ICA algorithm in electroencephalography [19]:
- -
-
the sources must be independent (signals come from statistically independent
brain processes);

- -
- there is no delay in signal propagation from the sources to the detectors (conducting media without delays at source frequencies);
- -
- the mixture is linear (Poisson's equation is linear);
- -
- the number of independent signal sources does not exceed the number of electrodes (we expect to have fewer strong sources than our 32 electrodes).
![]()
where ![]() is the so-called ``mixing'' matrix and each row of
is the so-called ``mixing'' matrix and each row of ![]() is a source's time activation. What we would like to find is an ``unmixing''
matrix
is a source's time activation. What we would like to find is an ``unmixing''
matrix ![]() , such that:
, such that:
![]()
or, in other words, ![]() ; but we do not know M, the only data we have is the
; but we do not know M, the only data we have is the ![]() matrix.
matrix.
Under the assumption of independent sources, ICA allows us to
construct such a ![]() matrix; however, since neither the matrix nor the sources are known,
matrix; however, since neither the matrix nor the sources are known, ![]() can be restored only up to scaling and permutations (i.e.,
can be restored only up to scaling and permutations (i.e., ![]() is not an identity matrix, but rather is equal to
is not an identity matrix, but rather is equal to ![]() , where
, where ![]() is a diagonal scaling matrix and
is a diagonal scaling matrix and ![]() is a permutation matrix). This problem is often referred to as Blind Source
Separation (BSS) [18, 31,
32, 33].
is a permutation matrix). This problem is often referred to as Blind Source
Separation (BSS) [18, 31,
32, 33].
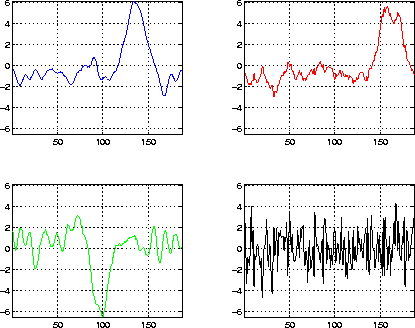
Figure 8: ICA activation maps obtained by unmixing the signals
from the signal subspace. We observe that there are only three independent
patterns, indicating the presence of only three separate signals in the
original data; the fourth component is noise.
The ICA process consists of two phases: the learning phase and the processing
phase. During the learning phase, the ICA algorithm finds a weighting matrix ![]() , which minimizes the mutual information between channels (variables),
i.e., makes output signals that are statistically independent in the sense
that the multivariate probability density function of the input signals
becomes equal to the product of
, which minimizes the mutual information between channels (variables),
i.e., makes output signals that are statistically independent in the sense
that the multivariate probability density function of the input signals
becomes equal to the product of ![]() probability density functions (p.d.f.) of every independent variable. This
is equivalent to maximizing the entropy of a non-linearly transformed vector
probability density functions (p.d.f.) of every independent variable. This
is equivalent to maximizing the entropy of a non-linearly transformed vector ![]() :
:
where ![]() is some non-linear function.
is some non-linear function.
There exist several different ways to estimate the W matrix.
For example, the Bell-Sejnowski infomax algorithm [18]
uses weights that are changed according to the entropy gradient. Below,
we use a modification of this rule as proposed by Amari, Cichocki and Yang
[20], which uses the natural gradient rather
than the absolute gradient of ![]() . This allows us to avoid computing matrix inverses and to speed up solution
convergence:
. This allows us to avoid computing matrix inverses and to speed up solution
convergence:
![]()
where the vector ![]() is defined as:
is defined as:
![]()
and for the nonlinear function g we used:
![]()
In the above equation ![]() is a learning rate and
is a learning rate and ![]() is the identity matrix [33]. The learning
rate decreases during the iterations and we stop when
is the identity matrix [33]. The learning
rate decreases during the iterations and we stop when ![]() becomes smaller than a pre-defined tolerance (e.g.,
becomes smaller than a pre-defined tolerance (e.g., ![]() ).
).
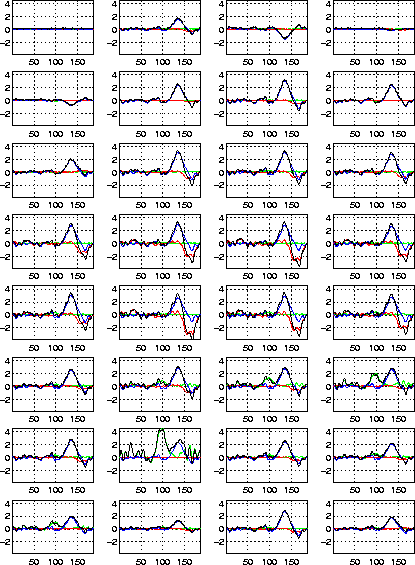
Figure: The projection of the first three activation maps from
Fig. 8 (as well as the original signals from
Fig. 7) onto the 32 electrodes.
The second phase of the ICA algorithm is the actual source separation.
Independent components (activations) can be computed by applying the unmixing
matrix ![]() to the signal subspace data:
to the signal subspace data:
Projection of independent activation maps ![]() back onto the electrodes one at a time can be done by:
back onto the electrodes one at a time can be done by:
![]()
where ![]() is the set of scalp potentials due to just the
is the set of scalp potentials due to just the ![]() source. For
source. For ![]() we zero out all rows but the
we zero out all rows but the ![]() ; that is, all but the
; that is, all but the ![]() source are ``turned off''. In practice we will not need the full time sequence
source are ``turned off''. In practice we will not need the full time sequence ![]() in order to localize source
in order to localize source ![]() , but rather simply a single instant of activation. For this purpose, we
set the
, but rather simply a single instant of activation. For this purpose, we
set the ![]() terms to be unit sources (i.e.,
terms to be unit sources (i.e., ![]() ), resulting in
), resulting in ![]() row elements which are simply the corresponding columns of
row elements which are simply the corresponding columns of ![]() .
.
Next: Source localization Up: Independent Component Analysis For Previous: Inverse Problem Zhukov Leonid
Fri Oct 8 13:55:47 MDT 1999