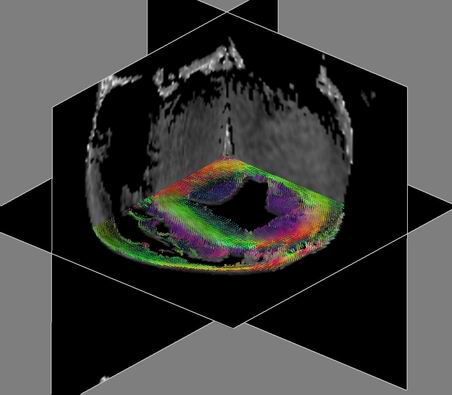
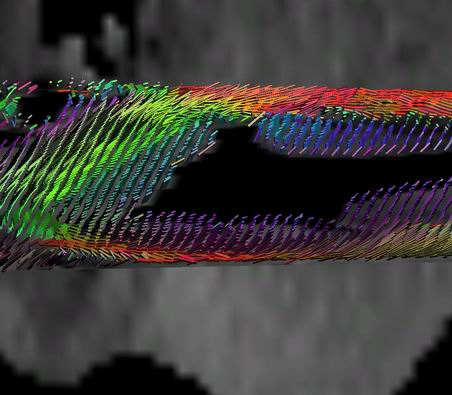
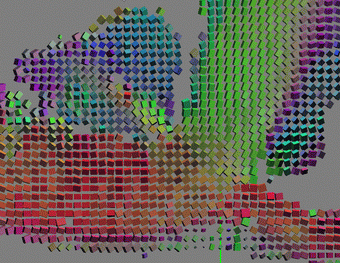| Figure 2:
Glyph-based visualization of the the raw data tensors. RGB colors
correspond to XYZ components of the largest eigenvector. Boxes are
oriented according to eigenvectors and scaled by eigenvalues. For example,
red boxes are oriented along the red (X) axis. Blue boxes correspond to
the vertical muscle fibers in the inside the ventricle. From left to
right: axial slice of the data; magnified region from the left image; the
same region with enhanced scaling for the glyph boxes emphasizing
principal direction. Note the transition from the red boxes along the X
axis to the green boxes, parallel to Y axis. This transition follows the
direction of the heart muscle fibers. |